MALDIPepQuant v3.1
User
manual (v1.0)
How to install MALDIPepQuantFirst of all, be sure to have at least version 1.5 of Java Runtime Environment (JRE) installed on your computer. If not, you can download the latest version from the Sun website. Download
latest version of the MALDIPepQuant distribution from here. |
How to start MALDIPepQuantDouble click on the
file called REPLACEBYVALUE inside the folder
MALDIPepQuant/exe/. |
How to run MALDIPepQuant
Fill out the fields according to the following informations and click on
the Start... button. 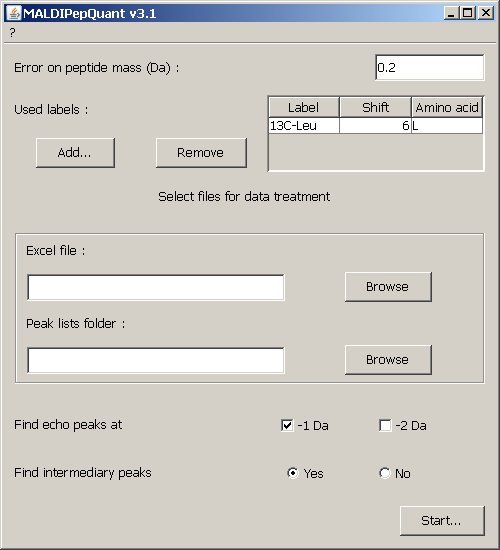 Label of heavy peptideThis is the label used to indicate the position of the heavy amino acid.ErrorAllowed error on the mass (Dalton).Amino acidHeavy aminoacid.ShiftWeight added to the heavy amino acid.DataExcel fileAn Excel export from Phenyx. See export named "Silac data export" in the Phenyx management console.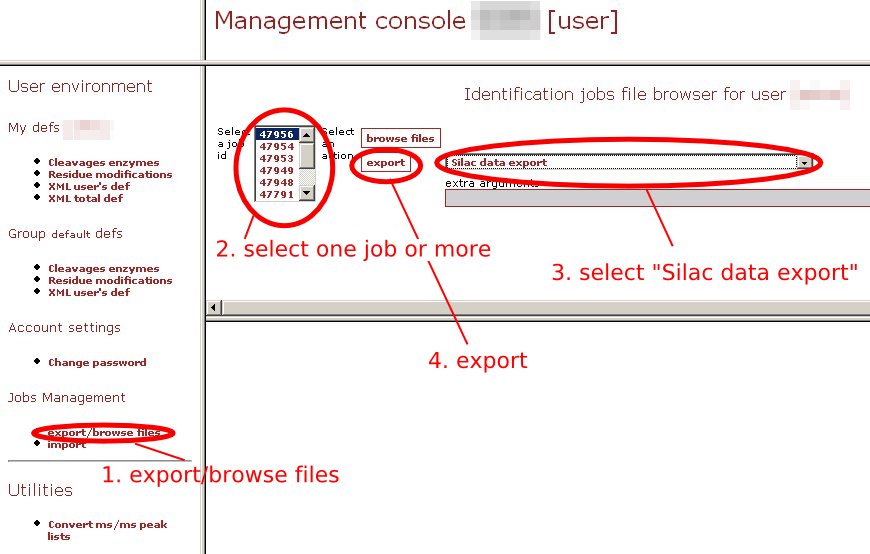
Peak Lists folderFolder containg all the MS peak lists in mgf format, as generated by a MALDI-TOF/TOF 4700 or 4800 from Applied Biosystems. Click on the Browse button, select one of the MS files and in the "File name" field replace the Label element by a #.For example if the name of the file was pmf_I14_118760352313.txt it will become pmf_#_118760352313.txt OptionsFind echo peaksSelect echo peaks you want to search. You can select either -1Da either -2Da, or both.Find intermediary peaksSelect this option if you think that the heavy amino acids were only partially incorporated. |
Content of the output fileOutput file will be created in the same folder as the original
Excel file (Phenyx export) with the "quanti_" prefix added to the
original file name. Columns detailsProteinsProtein of current peptide. Peptides Sequence of the peptide. Labeled AA nb Number of labeled amino acids in this peptide. Forms Observed forms for this peptide. Masses Labelof the MS spectrum followed by the masses found (light form/heavy form/eventual intermediary forms). Light Form Peak area found for the light form of the peptide. This column is followed by its eventual -1 and -2Da informations. Heavy Form Peak area found for the heavy form of the peptide. This column is followed by its eventual -1 and -2Da informations. Depending on the parameters, it will also be followed by the columns containing informations about the intermediary forms of the peptide. Sum LF Sum of the peak areas of the light forms of the peptide. Sum HF Sum of the peak areas of the heavy forms of the peptide (including eventual intermediary peaks). LF / HF Ratio of the sum of the light forms divided by he sum of heavy forms. |
