PeptideCutter - Special cleavage rules for trypsin and chymotrypsin
A general model of enzymatic cleavage:

Subsite nomenclature was adopted from a scheme created by Schechter and Berger (1967, 1968) and used in the following description of enzyme specificities. According to this model, amino acid residues in a substrate undergoing cleavage are designated P1,P2, P3, P4 etc. in the N-terminal direction from the cleaved bond. Likewise, the residues in C-terminal direction are designated P1', P2', P3', P4'. etc. as shown in Fig.1.
Establishing a specificity model for protease cleavage:
A first and obvious approach to obtain information concerning the cleavage specificity of a protease is the characterization of the respective natural substrate. In a next step, standard polypeptides can be used for digestion such as glucagon or insulin chains. However, an ideal polypeptidic substrate should contain all possible 400 combinations of dipeptide bonds, whereas in insulin and glucagon for example only small fraction of these combinations can be found (not to mention the possibilities of tetrapeptide composition when taking into account the sites P2 to P2'. Ideally, the cleavage data would be obtained by digesting all availble proteins in the databases, but this is beyond feasability. A more systematic and complete approach would be to test the proteases with substrates of low molecular weight such as di-, tri, tetrapeptides etc.
Unfortunately, the available data for most proteases in still very incomplete. Only for few proteases enough information has been accumulated that allows a statistical treatment (for details see Keil, 1992) resulting in a more complete and refined picture of cleavage specificity. This evaluation of the influence of the amino acid sequence on the potential cleavage sites should allow predictions of cleavage by proteolytic enzymes. In the case of chymotrypsin for example, 235 proteins were chosen containing 3136 cleavage sites ( Keil,1987). The statistical evaluation is based on certain assumptions and restrictions:
- The pool of data is sufficiently large.
- The probability of the occurrence of a specific dipeptide bond in a protein sequence is assumed to be proportional to the relative occurrence of the two amino acids taking part in the respective dipeptide bond. This is not true, as sequences are found to be formed non-randomly. The problem can be circumvented by choosing a sufficiently large collection of heterogeneous protein sequences.
- The statistical treatment does not take into account the reaction rates or percentage yields of the cleavage reactions. Only events of cleavage or non-cleavage are taken into consideration.
- Influences of the tertiary structure on protein cleavage are not taken into consideration.
The cleavage probabilities:
In the following, models of cleavage probabilities for trypsin and chymotrypsinare explained. These models are based on charts published by Keil, (1992).
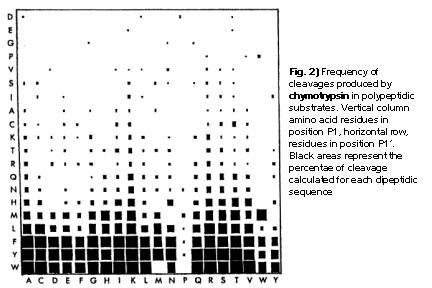
Chymotrypsin cleavage specificity:
A plot of P1 against P1' is presented in Fig.2. Here, the frequencey of cleavage for all 400 dipeptide sequences found in the above mentioned 235 proteins. To sum the results up, chymotrypsin preferentially cleaves at aromatic residues in position P1. It almost never cleaves at aspartic acid, glutaminic acid, glycine or proline. Certain amino acids have a favourable effect when positioned in P1, such as Lys or Arg. In contrast to that, Pro in P1' blocks almost all cleavage activities. The data used in the program PeptideCutter are derived from the size of the squares. In a simplified assumption that the largest value represent a cleavage probability of 100%, the other values are calculated in proportion to this.
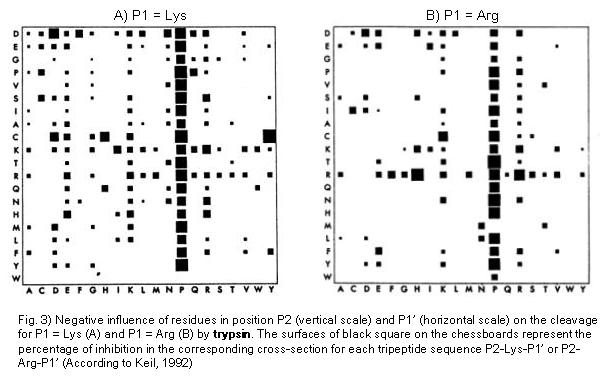
Trypsin cleavage specificity:
Trypsin preferentially cleaves at Arg or Lys in position P1. In a statistical study carried out by Keil (1992) the negative influences of residues surrounding the Arg- and Lys- bonds (i.e. the positions P2 and P1', respectively) during trypsin cleavage.The database LYSIS made it possible to access a pool of protein substrate data. The results of this study are presented in Fig. 3 A and B. Particularly interesting results are the following: Pro in Position P1' normally exerts a strong negative influence. Similarly, the positioning of R and K in P1' results in an inhibition, as well as negatively charged residues in positions P2 and P1'.
The PeptideCutter program does not take into consideration so-called "chymotrypsin-like" cleavages. These kind of cleavage events at aromatic or hydropohobic residues and are often reported (Keil , 1986). Nevertheless, they were attributed to impurities caused by traces of chymotrypsin and pseudotrypsin, a degradation product of trypsin.