Sulfinator - Documentation
The Sulfinator is a software tool able to predict tyrosine sulfation sites in protein sequences. It employs four different Hidden Markov Models that were built to recognise sulfated tyrosine residues located N-terminally, within sequence windows of more than 25
amino acids and C-terminally, as well as sulfated tyrosines clustered within 25 amino acid windows, respectively. All four HMMs contain the distilled information from one multiple sequence alignment. [More
on the data sets used to train and test the HMM]
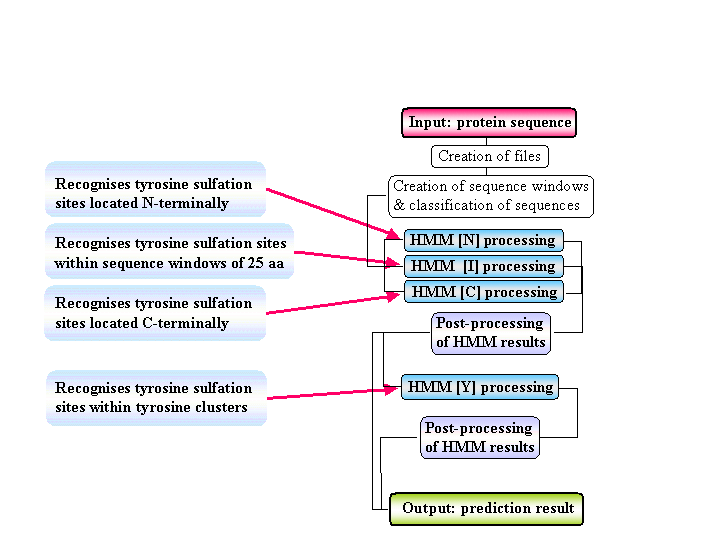 Fig: sulfinator flow chart
Fig: sulfinator flow chart
Protein sequences may be entered in either raw format or FASTA format. Tests of Swiss-Prot/TrEMBL sequences can also be performed by entering the protein identification (ID; eg. FA8_HUMAN) or accession number (AC; eg. P01050).
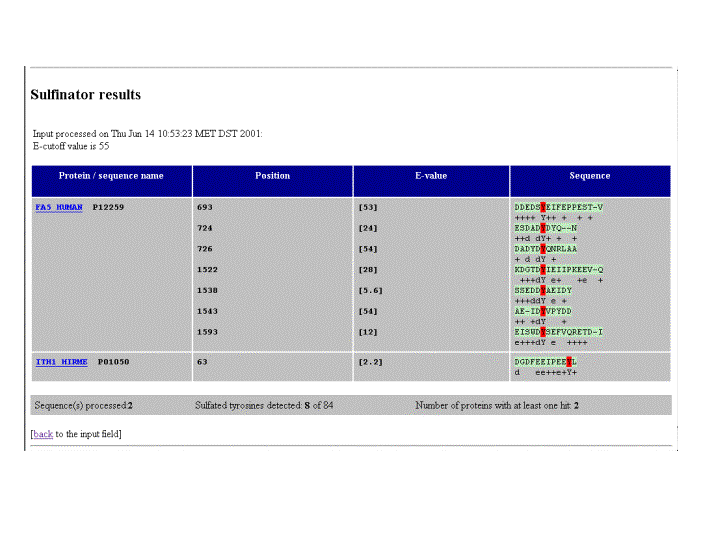
The output is an HTML table with 4 columns.
The first field contains the name (either entered by user or in case of raw format submission the first submitted sequence will be called "UNKNOWN1", followed by "UNKNOWN2" etc.) of the parsed sequence. The name will be hyperlinked to the corresponding Swiss-Prot/TrEMBL entry, in case a protein ID or AC number was used. In any other case, the name appearing in the first column is hyperlinked to a BLAST service against the Swiss-Prot/TrEMBL databases. The second column lists the position(s) of potentially sulfated tyrosines, and the third column gives a statistical value of the match. The fourth column shows where the entered protein sequence shows matches to the HMM chain (letters indicated positions where "exact" matches are located, capital letters mean highly conserved residues, and "+" marks positions with a positive score).
Monigatti F., Gasteiger E., Bairoch A., Jung E.
The Sulfinator: predicting tyrosine sulfation sites in protein sequences
Bioinformatics 18:769-770(2002).
PubMed: 12050077.
 Fig: sulfinator flow chart
Fig: sulfinator flow chart
Sequence input - supported formats:
Protein sequences may be entered in either raw format or FASTA format. Tests of Swiss-Prot/TrEMBL sequences can also be performed by entering the protein identification (ID; eg. FA8_HUMAN) or accession number (AC; eg. P01050).
Sulfinator output - results
The output is an HTML table with 4 columns.
The first field contains the name (either entered by user or in case of raw format submission the first submitted sequence will be called "UNKNOWN1", followed by "UNKNOWN2" etc.) of the parsed sequence. The name will be hyperlinked to the corresponding Swiss-Prot/TrEMBL entry, in case a protein ID or AC number was used. In any other case, the name appearing in the first column is hyperlinked to a BLAST service against the Swiss-Prot/TrEMBL databases. The second column lists the position(s) of potentially sulfated tyrosines, and the third column gives a statistical value of the match. The fourth column shows where the entered protein sequence shows matches to the HMM chain (letters indicated positions where "exact" matches are located, capital letters mean highly conserved residues, and "+" marks positions with a positive score).

For publication of results please cite:
Monigatti F., Gasteiger E., Bairoch A., Jung E.
The Sulfinator: predicting tyrosine sulfation sites in protein sequences
Bioinformatics 18:769-770(2002).
PubMed: 12050077.
